GeneCoder Molecular Biology Software
GeneCoder is a comprehensive and customizable Molecular Biology Software package for use in molecular cloning and DNA/protein sequence analysis. GeneCoder enhances the workflow of molecular cloning by allowing quick and easy sequence analysis and manipulation, allowing scientists to focus on the experiment.
GeneCoder 4.0 free for academic use only.
Download GeneCoder 4
(free for academic use)
GeneCoder 4 Features
Enhanced User Interface
- The new user interface introduced in GeneCoder v4 combines several features into a single window. Access sequences, tools, and view multiple sequences rather than hunting through multiple open windows.
- Quickly access several tools and features through right click popup menus.
- View sequences in multiple ways, from a sequence overview, to a plasmid map, to a digest panel and customize these views via the Plugin API.
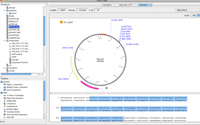
Sequence Database
- Organize sequences into one or more sequence repositories, and access sequences on the fly through the sequence database panel.
- Analyze one or more files directly through the database hierarchy popup menu.
- Construct sequence lists to compile or perform analysis on multiple sequences.
- Search all sequence files in the database by name, region, title, sequence, and/or much more...
Toolbox
- Access sequence analysis and manipulation tools on the fly through the toolbox panel
- Dynamically add new tools using the GeneCoder Plugin API.
Plugin API
- Write your own molecular biology tools using the GeneCoder plugin API.
Comprehensive Preferences
- Change preferences for restriction enzyme usage, preferred view component, region color, user interface look and feel, and much more...
Restriction Enzyme Analysis
- Dynamic Restriction Site Lists.
- Editable Restriction Enzyme Library.
- Restriction Enzyme Digests.
- Dynamic Restriction Maps.
Regions
- Highlight regions of interest in DNA and protein sequences, and include these regions in graphic representations such as restriction maps.
Alignments
- Pairwise alignments for both DNA and protein sequences.
- Remote access to the NCBI BLAST server to perform comprehensive BLAST sequence analysis.
- Search for common sequences from an editable database of sequences.
Files
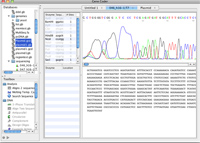
- Reads a variety of sequence file formats, including FASTA, Genbank, EMBL, ABI, DNA Strider, Text, and more...
- View, analyze, and import DNA sequence chromatogram traces.
- Export DNA and protein sequences into multiple file formats.
- GeneCoder uses the Genbank standard file format as the default file format.
Other Tools
- Calculate Oligonucleotide properties, including Melting Temperature, %GC content, and molecular mass.
- Translate DNA sequences into a protein sequence by one phase, two phase, and three phase.
- Perform remote Entrez Sequence Queries and download DNA and protein sequences from NCBI.
- Search for short sequences within a sequence file using text or regular expressions.


