GeneCoder - Command Line Utilities
Articles —> GeneCoder - Command Line Utilities
GeneCoder molecular biology software (v4.1) provides a route to automate its tools through the command line interface. Rather than running the software through the standard graphical user interface: the command line can be used to script and or otherwise access GeneCoder's tools. Launching the command line interface depends upon your operating system:
- Both linux and mac distributions come with a shell script named genecoder within the install director
- Windows users should install cygwin. Once installed, launch the cygwin terminal and 'cd' into the install directory (/cygdrive/c/Program Files (x86)/GeneCoder 4.0). To run the command line interface, launch the GeneCoder.exe file with the command line parameter '-cl'
Launching the utilities with no parameters will prompt a list of options available. Current default options include:
- restrictionsites: calculate restriction sites of a sequence
- malign: performs a multisequence alignment
- entrez: Runs eSearch and eFetch through NCBI
- translate: translates the input sequence(s)
- blast: blasts the sequence(s) through NCBI
- antiparallel: antiparallels the input sequence(s)
- pairwise: pairwise aligns the input sequences
- mutate: mutates a given sequence
- convert: file format conversion
Each tool has its own unique command line parameters. This being said, most tools will take input from the standard input (stdin) or from a file (typically specified via the -i command line parameter). For instance, on linux/mac one can cat a sequence file and pipe (|) the contents into GeneCoder. Default output is to the command line, however most tools will accept a -o parameter specifying the path to the output file.
As an example, one can query NCBI for a particular sequence:
./genecoder entrez -db protein -max 2 -q Csn5 -rm text -rt gb
The above results in a genbank text formatted results for the protein Csn5, retrieving the maximum of 2 records. Note that long sequences may take some time to download, and multiple queries should respect NCBI's policy on automated queries.
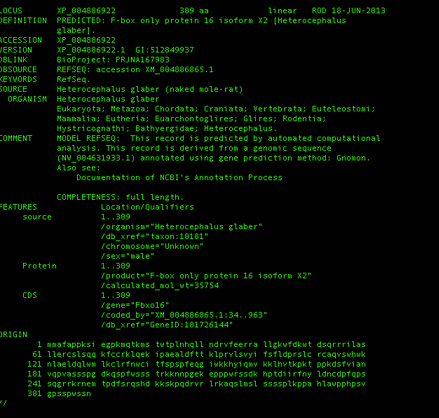
Command line search of NCBI
Note that all current command line tools are written through the Plugin interface, and new tool capability is available for those with knowledge of java through the Plugin API.
There are no comments on this article.